About
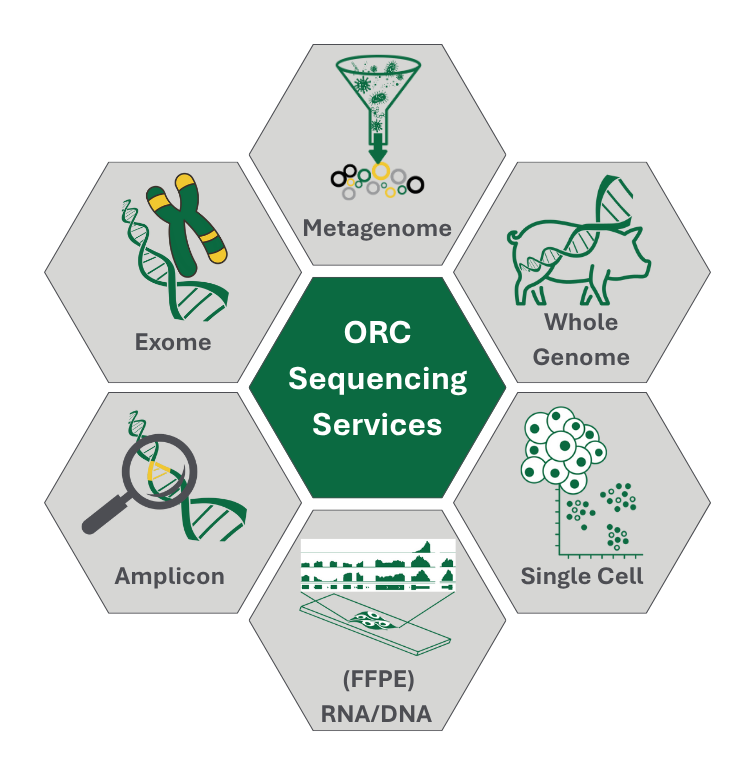
Welcome to the Omics Resource Centre (ORC)!
ORC is a next-generation sequencing (NGS) service laboratory and research support core operating in the Western College of Veterinary Medicine (WCVM) at the University of Saskatchewan (USask) in Saskatoon, Sask.
ORC provides integrated genomics services to its various partners at USask, other academic and non-profit research institutions, and for-profit businesses or institutions.
In addition, the Omics Resource Centre serves as a training hub for highly qualified personnel in USask. ORC also offers consultation services — from study design to data analysis — that are tailored to the needs of veterinary, animal agriculture and basic research communities.
Our mission
- To serve as a permanent, modular and sustainable omics infrastructure performing crude sample processing and nucleic acid extraction, NGS library preparation and sequencing services.
- To function as an extension of the academic research environment, tailored to meet evolving scientific needs with precision, scalability and professional-grade execution by offering custom standard operating procedure (SOP) development with and without the goal of automation.
- To support genomics and multiomics research across veterinary medicine, animal science, environmental stewardship, and related disciplines with consultations, instrument access and training for highly qualified personnel (HQPs).
If you have any questions or want to obtain a service quote, please email orc@usask.ca.
Visit the Consultation section to book a free project consultation with the ORC Lab Manager.
For sample submissions, click here to enter the Customer Portal.
Click here to view our Terms and Conditions.
Services
ORC offers a comprehensive menu of services including:
- Next Generation Sequencing
- Crude sample processing and nucleic acid extraction services
- DNA/RNA Quantification and Library QC
- Manual and high-throughput NGS library preparation
- Paid and piggyback training opportunities for USask highly qualified personnel (HQPs)
- Custom standard operating procedure (SOP) development
- Instrument booking
- Consultations
All services are supported by standardized operating procedures (SOPs), which are under continuous development and refinement in collaboration with researchers. As workflows evolve, ORC updates its offerings to remain at the cutting edge of genomics technology while maintaining a commitment to transparency and quality control.
ORC is also equipped to scale operations from individual pilot samples to full cohort studies, accommodating both exploratory and high-throughput research needs. Automation is central to this scalability, with core equipment including robotic liquid handlers, nucleic acid extraction systems, and modern short-read sequencers — all co-ordinated via an integrated Laboratory Information Management System (access ORC LIMS here) with ELN functionality and a Customer Portal.
Follow the link here to learn more about sample requirements and click here to book a project consultation.
Sequencing is performed using state-of-the-art AVITI Sequencer from Element Biosciences. We deliver sequencing data with high sequencing accuracy with Phred quality score (Q score) of above 40. ORC’s sequencing capabilities are designed to serve a wide range of applications, including (meta) genomics and (meta/single cell) transcriptomics. ORC customers will be provided with (1) demultiplexed FASTQ files, (2) Index Assignment file, (3) run metrics overview, (4) in-depth data QC, and (5) Run Manifest. ORC offers also an optional adapter trimming service (free-of-charge) and guidance for secondary NGS data analysis (on demand, click here to book a consultation).
ORC offers a growing catalogue of sequencing services based on proven and validated workflows. These currently include:
- RNA-Sequencing (3’mRNA, total RNA, mRNA, small RNA-Seq, and others)
- DNA Sequencing (Whole genome sequencing (WGS), low pass WGS, amplicon, exome, …)
- 16S/18S/ITS sequencing
- Metagenome sequencing
- Metatranscriptome sequencing (MetaT-Seq)
- Stowaway sequencing runs in small pilot projects
- Direct sequencing of customer-supplied libraries
RNA-Sequencing (RNA-Seq) refers to the starting material as all the libraries end up as DNA for AVITI sequencing. We offer RNA-Seq library preparation, with multiple options (fees apply) such as host RNA removal (for Metatranscriptome-Seq), ribodepletion, mRNA enrichment, 3'mRNA-Seq, and small RNA library preps. All our library preps are strand specific (for more information follow the link here). The term "RNA-seq" is used for protocols that sequence the entire transcripts; in contrast 3'mRNA-Seq refers to a low-cost protocol for differential gene expression (DGE) profiling that only sequences one piece at the 3'-end of a transcript. ORC customers are getting free access to a web-based interactive RNA-Seq analysis platform for complimentary data analysis.
More RNA-Seq workflows will become available soon (e.g. SLAM-Seq and single-cell RNA-Seq) and will be announced on the ORC website.
DNA-Sequencing refers specifically to high-pass (hp)- and low-pass (lp)-whole genome sequencing (WGS). The typical hp-WGS refers to the process of sequencing an individual's genome at a standard depth of 30x to 50x coverage for a high-resolution, base-by-base view of the genome that captures both large and small variants and delivers large amounts of sequencing data to support assemblies of novel genomes. ORC is capable of simultaneously sequencing either up to 6 human genomes at a 30-fold coverage or up to 100 metagenome samples with a coverage of 20 M 150bp paired-end reads/sample.
In contrast, lp-WGS is a cost-effective method that sequences an entire genome at a low depth (typically between 0.3-to-5-fold coverage), making it cheaper and faster than hp-WGS (e.g. 400 human samples at a 0.5-fold coverage). This method is useful for detecting large-scale variations like copy number variations (CNVs) and small variations like single nucleotide polymorphisms (SNPs) in population studies. LpWGS offers a broad genomic overview with greater statistical power for applications like genome-wide association studies (GWAS) compared to traditional genotyping arrays or reduced representation libraries for e.g. ddRADSeq.
Amplicon-Sequencing is a targeted DNA sequencing method that provides an opportunity for the interrogation of specific regions of the genome to obtain information about the genetic regions of interest (ROI). In amplicon sequencing, highly multiplexed PCR generates amplicons of targeted regions of genomic DNA, after which adapters are added via PCR using indexing primers with either combinatorial dual indexes (CDIs) or unique dual indexes (UDIs). The final library fragments are pooled and sequenced.
This technique is more cost-effective and faster than whole-genome sequencing, making it ideal for identifying genetic variants, studying microbial communities, and tracking pathogens.
Our standard amplicon sequencing workflow in phylogenetic studies utilizes highly efficient commercial kits for prokaryotic 16S and eukaryotic Internal transcribed spacer (ITS) ribosomal RNA (rRNA) sequencing library preparation.
The prokaryotic 16S rRNA gene is approximately 1500 bp long and contains 9 variable regions (V1-V9) interspaced by conserved regions. Specific contiguous and non-contiguous combinations of these regions show better classification accuracy across different environmental niches (click the link here form more information). We typically utilize NGS library preparation kits targeting either the V1-V3 region for higher species-level resolution for specific communities, or the V3-V4 region for broader taxonomic coverage, good genus-level resolution, and suitability for high-throughput studies.
ITS rRNA gene sequencing is a routine technique for mycobiome (fungal) composition profiling. Compared to shotgun metagenomics sequencing, ITS rRNA gene sequencing is less impacted by the presence of non-microbial DNA, more cost-effective and more robust requiring less input DNA. We utilize NGS library preparation kits targeting the ITS3/ITS4 primer sets which showed superior performance (specificity, higher proportion of fungal sequences) over other ITS regions and 18S rRNA gene amplicon sequencing (click the link here and here for more information).
ORC also assists with the design of custom amplicon panels. Follow the link here to book a project consultation.
Metagenome-Sequencing is an untargeted method that provides access to the full genetic content in a sample and is therefore ideal for microbial community profiling of samples from different natural environments, for example in identification of new pathogens, identify and track genomic mutations, or assemble whole genomes.
ORC utilizes automated protocols to extract microbial DNA with and without optional host DNA-depletion from a variety of solid and liquid samples including body fluids. We typically use an enzymatic NGS library preparation protocol that includes PCR-free and PCR-plus options and generates a linear library that is circularized onboard the AVITI sequencer. Follow the link here for more information.
Metatranscriptome-Sequencing (MetaT-Seq) is a more laborious “shotgun” sequencing technique of reverse transcribed microbial RNA used to study gene expression in native microbial communities without the need for cultivation. MetaT-Seq can be used for the identification of novel genes, functional characterization of active bacteria, and community metabolic interaction in microbial ecology research.
ORC employs an automated bead-based extraction kit in combination with the Thermo Scientific KingFisher™ Apex purification system for the unbiased high-throughput isolation of RNA from microbes including gram positive/negative bacteria, fungi, protozoans, and viruses from almost any sample type. The depletion of microbial ribosomal RNA is included in the basic service. Optionally, ORC customers can choose to enrich the desired microbial RNA by removal of any host RNA and/or enrichment of bacterial mRNA (fees apply). Follow the link here for more information.
Direct sequencing of pre-pooled NGS libraries is offered with and without customer-supplied NGS sequencing kits.
The vast majority of third-party NGS libraries are compatible with ORCs AVITI sequencer. Check for incompatible assays and workflows following the link here. Follow the link here to book a project consultation and contact ORC via email (orc@usask.ca) to request an estimate.
ORC offers DNA and RNA extraction services from almost any liquid or solid crude sample from animals, plants, environments, pathogens, and cell cultures including formalin-fixed paraffin-embedded (FFPE) samples or samples in nucleic acid (NA) stabilization solutions (e.g. Invitrogen RNAlater™ or Zymo Research DNA/RNA Shield™). A prerequisite to offer this service is the samples to be free of active pathogens. Therefore customers are required to inactivate all pathogens prior to sample submission to ORC and provide the critical sample information in the Sample Submission Template or the Customer Portal.
ORC employs (semi-) automated extraction kits in combination with the Thermo Scientific KingFisher™ Apex purification system to enable high-throughput nucleic acid isolation for a wide range of applications.
Crude sample extractions can be booked as stand-alone service or in combination with quality control and quantification workup or sample normalization and aliquoting.
Alternatively, crude sample extraction services are available as part of a full end-to-end service including NGS library preparation, NGS library QC and AVITI sequencing.
On customer request we can modify the workflow-specific standard crude sample processing to include addon features such as host NA depletion, ribosomal RNA depletion or simultaneous extraction of DNA and RNA from the same sample, if applicable.
Please refer to the ORC Sample Submission Guideline for more information on specific sample requirements.
Quantification and quality control (QC) of nucleic acid (NA) extract and NGS libraries are essential prior to sequencing. Our NA and library preparation services include QC on the Tapestation (Agilent Technologies), and quantification using the Qubit Flex Fluorometer (Life Technologies) and/or the Synergy LX microplate reader (Agilent Technologies). In some instances (e.g. PCR-free libraries prior to pooling or endpoint PCR cycle number optimization) a higher level of precision is required, and we use either the KAPA Biosystems SYBR FAST Library Quantification Kit for Illumina Sequencing Platforms or the Lexogen PCR Add-on and Reamplification Kit V2 for Illumina to assess the quantities.
We offer also stand-alone services for NA and library quantification and a variety of Tapestation QC assays (RNA, DNA D5000, DNA D1000, genomic DNA, standard or high sensitivity). Alternatively, ORC customers can perform sample quantification and QC on their own by booking the instruments and accompanied customer training service for a fee using the ORC LIMS or by sending a request via email to orc@usask.ca.
If you would like us to pool your libraries for direct sequencing on the AVITI sequencer, we can do this too although we prefer the submission of pooled libraries.
For more details on the sample submission requirements, please refer to the ORC Sample Submission Guideline.
Please review the Pricing section of for detailed pricing information.
Next-Generation Sequencing (NGS) builds up on the existence of a large diversity of library creation methods tailored to different experimental approaches, with numerous commercial kits and variations for each method.
ORC offers common library prep methods as “standard” services with a fixed price and established expectations for turnaround and performance. ORC employs liquid handling robots to minimize sample handling variation and to provide fast turnaround times. Our library preparation services include library QC, library quantification, and library pooling. All our libraries are barcoded (single indexed or uniquely-dual-indexed). The typical insert size of our libraries is kit-specific and ranges between 250bp and 900bp.
Successful sequencing is dependent not only on the quality of submitted DNA or RNA but also on the quality of the library created and in the selection of the appropriate library kit. Our team will guide you through the selection process during the project consultation to ensure the highest NGS library quality and maximal data output of your flow cell. We are looking forward to discussing the individual options – including custom or specialized libraries - and protocols suitable for your specific research projects.
We currently offer:
- Genomic DNA libraries (whole-genome shotgun libraries, metagenome libraries)
- RNA-Seq libraries (3’mRNA, total RNA, mRNA, small RNA-Seq, FFPE RNA-Seq, metatranscriptome)
- Amplicon libraries (16S/18S/ITS, custom)
- High-Throughput (HT) library preps
- PCR-free libraries
- Custom libraries (on demand)
Aside from offering sequencing services ORC also acts as a training hub for highly qualified USask personnel (HQP). On demand and where operational capacity allows, students, postdocs, research technicians, and research associates may be trained to use specific instruments or workflows. The training is offered in two ways:
- Piggyback Training: HQPs receive training by shadowing ongoing ORC-run service projects (free-of-charge).
- Paid Individual Training: Users can book one-on-one training sessions with ORC staff to learn specific workflows or instrument operation (fees apply, visit the Pricing section).
If you have any questions or want to obtain a service quote, please email orc@usask.ca.
ORC offers instrument access bookings for USask researchers and highly qualified personnel needing specific equipment but lacking the infrastructure in their own labs. Instruments currently available for booking (fees apply) include:
- Agilent Tapestation 4200
- Agilent Synergy LX Microplate reader
- KingFisher Apex Nucleic Acid Purification System
- Invitrogen™ Countess™ 3 FL Automated Cell Counter
- Miltenyi Biotec gentleMACS™ Dissociator
USask researchers and HQPs using ORC equipment must be trained as a fee for service by ORC staff. Once trained, USask members can get access to the facility and book the instrument whenever available (fees apply).
Prior to approval of instrument booking access first time users are required to read the ORC Biosafety Plan and the ORC General Procedures documents.
For more details, please refer to Pricing section and the Equipment Booking Guidelines.
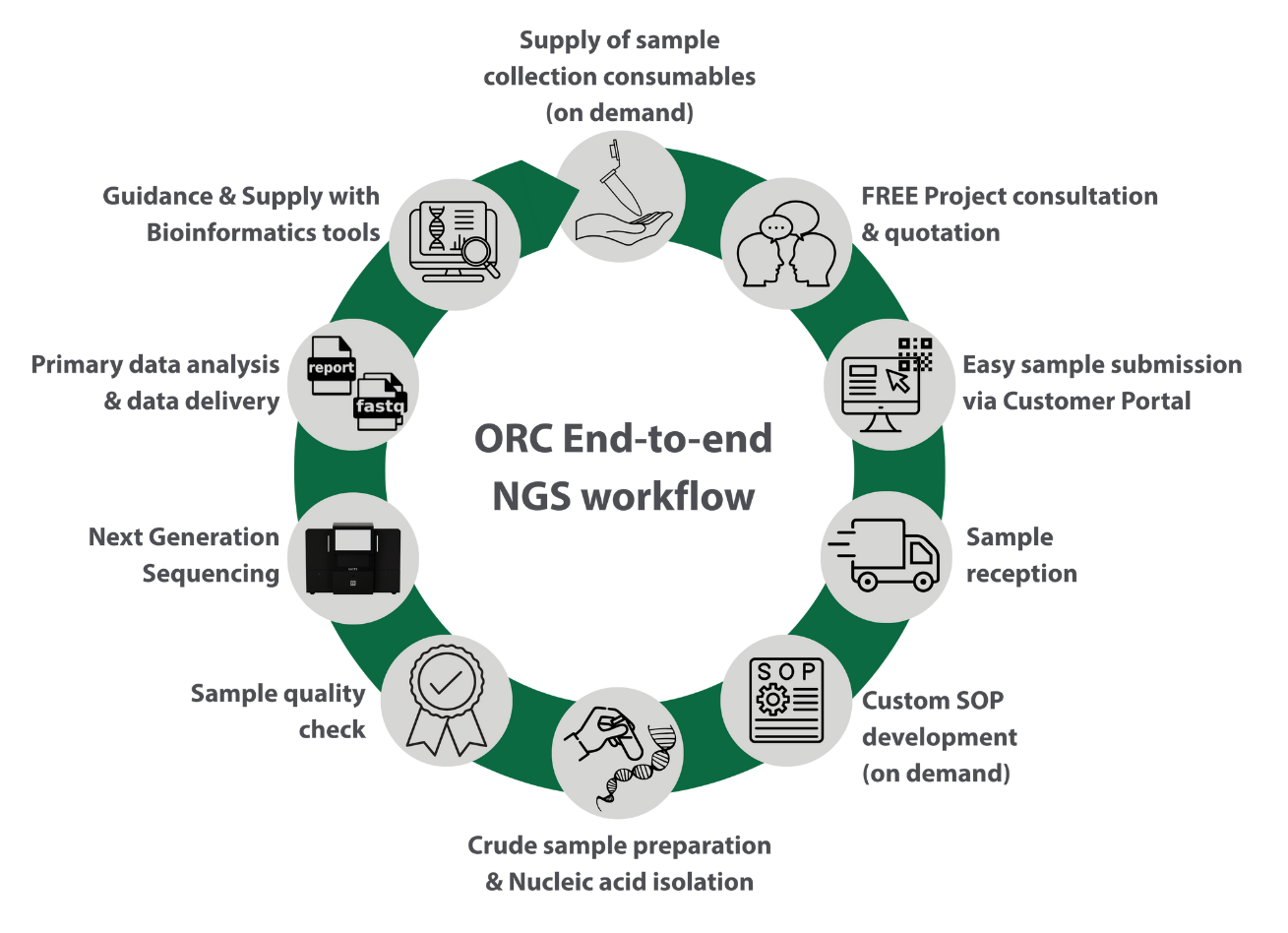
New customers are required to first reach out to orc@usask.ca in order to request a quote. Ideally, this happens after the customer had booked a free-of-charge consultation with the ORC lab manager in person or online via Teams. Click here to book a consultation.
After written acceptance of the quote by the customer, they receive access to the ORC Customer Portal for providing their contact and financial information, placing a service request and providing any required information related to their project and samples.
The ORC Lab Manager will review the service request and permit the sample submission based on the completeness of the information given.
Equipment & Platforms
ORC’s operations are physically and functionally separated across three WCVM locations.
- Sample receiving and storage (Room C616)
- Crude sample processing (Room 2259)
- NGS library preparation and sequencing (Room 2207)
Each space is equipped for specific workflow stages, with automation integrated where practical. Examples include robotic liquid handlers, automated purification systems and modular library preparation stations. This separation supports traceability, minimizes contamination and aligns with ORC’s principle of seamless, high-integrity processing.
ORC provides the latest NGS high-end benchtop platform from Element Biosciences — the AVITI sequencer. This powerful tool enables researchers to pursue a wide range of sequencing projects for any budget and scope.
All systems, workstations and storage locations at ORC are integrated into a laboratory information management system (LIMS) to ensure effective management of samples, projects and associated information or workflows.
A selection of ORC’s core equipment and those that can be booked on a per-use basis are listed here. For more details on equipment bookings, visit the Pricing section and the ORC Equipment Booking Guideline.

- The Omics Resource Centre (ORC) operates the Element Biosciences AVITI System, a short-read, high-end benchtop sequencer that delivers a lower cost per Giga base pair (Gbp) without the need to scale throughput. This instrument features two independently operating flow cells and up to four individually addressable lanes to give ORC a greater sequencing flexibility while maintaining the option to deliver up to 2 billion reads of combined sequencing output. The AVITI delivers the most accurate short-read sequencing available today, with most bases scoring above a Phred scale Q-score of 40 and partially above 50.
- AVITIs higher sequencing accuracy is achieved by a novel sequencing technology, termed avidite base chemistry (ABC), that reduces run costs through reduced concentration of reagents and improves performance through improved base detection and high-fidelity amplification without index hopping artifacts. The AVITI readily adapts to a variety of applications, offering methods that scale from amplicons to whole genomes. With outstanding data quality and compatibility with existing library formats, the AVITI will be of particular interest to single-cell and spatial researchers, as well as researchers who require ultra-high-quality reads. Click here to learn more about the Element AVITI system.
- Adept Workflow for third party (mostly linear) libraries — circularizes linear libraries prepared with a compatible third-party library prep kit (i.e. Illumina) to allow researchers to continue using the same library prep and analysis tools.
- Elevate Workflow for native prep — prepares dual-indexed linear libraries from genomic DNA input and libraries are circularized on the flow cell as part of the sequencing run. Include the choice of PCR-free or PCR-plus.
- The AVITI system is not available for instrument booking.

- The Omics Resource Centre (ORC) crude sample extraction suite hinges on the KingFisher Apex Purification System for (semi)-automated high and low volume throughput isolation of high-quality nuclei acids, proteins and cells from any specimen.
- Using a simple process (bind, wash, elute), the KingFisher Apex utilizes an effective system of permanent magnetic rods with disposable tip combs to collect and transfer beads from solution instead of moving the liquids to produce highest sample purity and integrity for high-end downstream applications such as qPCR, NGS, digital PCR, and mass spectrometry
- Depending on the particular chemistry employed and protocol modifications nucleic acids from 1 to 96 samples can be extracted in as little as 1 hour. Click here to learn more about the KingFisher Apex Purification System.
The KingFisher Apex Purification System is available for instrument booking. For first-time users, a one-hour hands-on training session is required to be booked in addition. Visit "Pricing" for more information on training and booking fees.

- The Omics Resource Centre (ORC) offers high-throughput automation platforms for quality, quantity and integrity assessment for up to o 96 nucleic acid samples per run.
- Agilents 4200 TapeStation system offers high-throughput fully automated electrophoresis for quality and integrity control for up to 96 nucleic acid samples per run in less than 90 minutes. With as little as 1 to 2 µL of sample volume reliable integrity standards for RNA (RNA integrity number equivalent, RINe), genomic DNA (DNA integrity number, DIN), cell-free DNA (%cfDNA), and formalin-fixed paraffin-embedded (FFPE) samples (DV200) can be calculated for a variety of workflows such as NGS library QC, DNA/RNA-based vaccine QC and biobanking.
- The TapeStation System is available for instrument booking. For first-time users, a one-hour hands-on training session is required to be booked in addition. Please visit “Pricing" for more information on training and booking fees.

- The Omics Resource Centre (ORC) offers high-throughput automation platforms for quality, quantity and integrity assessment for up to 96 nucleic acid samples per run.
- With continuous wavelength selection for UV-Vis measurements ranging from 200 to 999 nm in 1 nm increments and filters for fluorescence and luminescence the Agilent BioTek Synergy LX automates many common end-point assays, including nucleic acid and protein quantification, ELISA, BCA, and Bradford assay, and cell viability assays for 1 to 384 samples simultaneously.
- The BioTek Synergy LX Multimode Reader is available for instrument booking. For first-time users, a one-hour hands-on training session is required to be booked in addition. Visit “Pricing" for more information on training and booking fees.

- The Omics Resource Centre (ORC) integrates NA purification and NGS library preparation workflows for manual low throughput and automated high throughput demands.
- The Microlab® STAR Liquid Handler offers flexible customization, able to meet the needs of any automation application or requirement. For example, ORCs Microlab® STAR is specifically configured for automated NGS library preparation with an On-Deck Thermal Cycler (ODTC), several Hamilton Heater Shaker (HHS), a barcode reader, and two Inheco CPAC (cold plate air cooled) heater/coolers. The configuration enables demanding automation workflows with longer walkaway times, autoloading of samples and consumables, and cooling of sensitive reagents and libraries.
- Up to 96 samples with volumes between 1 and 5000ul can be processed and complex protocols requiring up to 45 ANSI plates can be implemented.
- The Microlab® STAR Liquid Handler is not available for instrument booking.

- The Omics Resource Centre (ORC) integrates NA purification and NGS library preparation workflows for manual low throughput and automated high throughput demands.
- The epMotion® 5073 liquid handler is part of ORCs crude sample extraction suite and enables automation for medium throughput pipetting tasks such as PCR and qPCR applications, DNA/RNA normalization, cherry picking, dilution series and reformatting.
- Up to 96 samples with volumes between 0.2 and 1000ul can be processed and protocols requiring up to 6 standard microplates can be implemented.
- The epMotion® 5073 Liquid Handler is not available for instrument booking.

- Aside from high-throughput automation platforms the Omics Resource Centre (ORC) integrates low- to medium throughput platforms for quality and quantity assessment such as the Invitrogen™ Qubit™ Flex benchtop fluorometer.
- The Qubit™ Flex is part of ORCs NGS core lab and enables highly accurate quantification of DNA, RNA, microRNA, and protein for quantitation, next-generation sequencing, and endotoxin detection. The Qubit™ Flex measures fluorescence of up to eight samples simultaneously.
- The Qubit™ Flex comes with on-board features such as ‘Assay Range Calculator’ to help you determine which Qubit™ assay provides the most accurate quantification based on your sample volume and estimated sample concentration, and a ‘Normalization Calculator’ that helps you to normalize your samples of variable concentration to the same molarity, concentration, or mass using the results from your assay.
- The Qubit™ Flex Fluorometer is not available for instrument booking.

- The Omics Resource Centre (ORC) is offering single cell RNA sequencing (scRNA-Seq) workflows from nucleic acid extraction to analysis of transcriptomic heterogeneity at the single-cell level to facilitate new insights into cellular responses in research areas such as cancer research, cell development and function, and immunology.
- The Countess 3 FL counter allows brightfield or fluorescence cell counting with three-channel flexibility (brightfield and two optional fluorescence channels), enabling researchers to count cells and isolated nuclei, monitor fluorescence, examine apoptosis, and measure cell viability.
- The Countess™ 3 FL Automated Cell Counter is available for instrument booking. For first-time users, a one-hour hands-on training session is required to be booked in addition. Visit "Pricing" for more information on training and booking fees.

- The Omics Resource Centre (ORC) is offering single cell RNA sequencing (scRNA-Seq) workflows from nucleic acid extraction to analysis of transcriptomic heterogeneity at the single-cell level to facilitate new insights into cellular responses in research areas such as cancer research, cell development and function, and immunology.
- The gentleMACS™ Dissociator is a benchtop instrument for the semi-automated dissociation of tissues into single-cell suspensions or thorough homogenates. One or two samples can be processed at a time using proprietary gentleMACS Tubes with the instrument. The instrument offers optimized gentleMACS tissue homogenization programs for a variety of specific applications (e.g. single-cell suspensions; total RNA, protein or mitochondria isolation; viral/bacterial load determination) and tissues (heart, spleen, lung, muscle tumor, neural, skin). Processing time can span from a few minutes to overnight assays.
- The gentleMACS™ Dissociator is available for instrument booking. For first-time users, a one-hour hands-on training session is required to be booked in addition. Visit "Pricing" for more information on training and booking fees.
Pricing
The Omics Resource Centre (ORC) provides services at three recharge rate scales.
- USask rates applies to projects paid from USask accounts. The Goods and Service Tax (GST) will not be applied to this rate.
- Academic rates apply to all other academic and non-profit research, including government institutions on and off-campus (such as Agriculture and Agri-Food Canada, National Research Council of Canada and others).
- Industry rates apply to projects from for-profit businesses or institutions. Taxes will apply for academic and industry rates.
Per sample prices listed below* are examples based on a bulk submission of 96 samples and will vary based on the actual sample number and the desired target coverage. Customers are required to request an individualized quote and a project consultation session before sample submission. Click here to book a consultation session.
More information on submission requirements can be found in the Terms and Conditions and Sample Submission Guideline. Please contact ORC (orc@usask.ca) for more details on fees for standard or custom services such as SOP development.
Note: pricing is subject to change. Please check regularly for any updates.
|
Service |
Assay |
USask* |
Academic* |
Industry* |
Comments |
|
Sample QC (i.e. size, integrity, purity) |
Tapestation (RNA, D5000, D1000, genomic, standard or high sensitivity) |
$10-$14 |
$11-$15 |
$14-$20 |
Customer required to provide purified nucleic acids, DNA size range between 35bp – 60kbp, detection limit RNA: 100pg/µl; DNA: 5pg/µl |
|
Sample quantification |
RNA, DNA, sscDNA, dscDNA, libraries |
$2 |
$2.50 |
$3 |
Customer required to provide purified nucleic acids, detection range RNA: 0.1–500ng/µl; DNA: 0.2-400ng/µl |
|
Ribodepletion (rRNA depletion, sample QC & quantification, QC report) |
mRNA enrichment |
$142 |
$158 |
$209 |
Can be booked as an add-on to service packages or as a stand-alone service. |
|
Direct sequencing (excluding sequencing kit, customer provided) |
Any |
$788-$1,779 |
$875-$1,976 |
$1,167-$2,635 |
Sequencing on submitted library pools. Customers are required to provide sample QC, index information, and the entire sequencing kit. Pricing depends on the sequencer run times. |
|
Direct sequencing (sequencing kit included) |
Any |
$1,915-$4,896 |
$2,128-$5,440 |
$2,837-$7,253 |
Sequencing on submitted library pools. Customers are required to provide sample QC and index information. Pricing depends on the sequencing kit and sequencer run times. |
|
Bundle: total RNA extraction + 3'mRNA-Seq (end-to-end workflow from crude sample to FASTQ data) |
3' Tag-Seq Gene Expression Profiling |
$153 |
$169 |
$220 |
Ribodepletion can be added as option. Free access to web-based interactive RNA-Seq analysis platform for complimentary data analysis included. |
|
Bundle: FFPE RNA extraction + 3'mRNA-Seq (end-to-end workflow from crude sample to FASTQ data) |
3' Tag-Seq Gene Expression Profiling |
$163 |
$179 |
$230 |
Ribodepletion can be added as option. Free access to web-based interactive RNA-Seq analysis platform for complimentary data analysis included. |
|
Bundle: total RNA extraction + Total RNA-Seq (end-to-end workflow from crude sample to FASTQ data) |
Full-length transcript analyses (isoforms, alternative splicing studies, lncRNA, Phusion transcript, variant analyses, and many more) |
$186 |
$204 |
$263 |
Ribodepletion can be added as option. Free access to web-based interactive RNA-Seq analysis platform for complimentary data analysis included. |
|
Bundle: total RNA extraction + mRNA-Seq (end-to-end workflow from crude sample to FASTQ data) |
Full-length transcript analyses (isoforms, alternative splicing studies, lncRNA, Phusion transcript, variant analyses, and many more) |
$195 |
$213 |
$276 |
Free access to web-based interactive RNA-Seq analysis platform for complimentary data analysis included. |
|
Bundle: FFPE RNA extraction + Total RNA-Seq (end-to-end workflow from crude sample to FASTQ data) |
Full-length transcript analyses (isoforms, alternative splicing studies, lncRNA, Phusion transcript, variant analyses, and many more) |
$176 |
$196 |
$203 |
Ribodepletion can be added as option. Free access to web-based interactive RNA-Seq analysis platform for complimentary data analysis included. |
|
Bundle: Microbial RNA extraction + Metatranscriptome-Seq (end-to-end workflow from crude sample to FASTQ data) |
Full-length transcript analyses (isoforms, lncRNA/sRNA studies, variant analyses, gene expression profiling, and many more) |
$278 |
$306 |
$398 |
Ribodepletion is standard and included in the pricing. Optional bacterial mRNA enrichment and host RNA removal procedures are available on demand. |
|
3'mRNA-Seq (sample QC, library prep & QC, sequencing, demultiplexed FASTQ data & QC report) |
3' Tag-Seq Gene Expression Profiling |
$134 |
$147 |
$190 |
Customers are required to submit, DNA-free RNA extracts and provide sample quantification & QC data. Low RNA quality samples are permitted. Free access to web-based interactive RNA-Seq analysis platform for complimentary data analysis included. |
|
mRNA-Seq (sample QC, library prep & QC, sequencing, demultiplexed FASTQ data & QC report) |
Full-length transcript analyses (isoforms, alternative splicing studies, lncRNA, Phusion transcript, variant analyses, and many more) |
$179 |
$196 |
$253 |
Customers are required to submit only high-quality, DNA-free RNA extracts and provide sample quantification & QC data. Free access to web-based interactive RNA-Seq analysis platform for complimentary data analysis included. |
|
Total RNA-Seq (sample QC, library prep & QC, sequencing, demultiplexed FASTQ data & QC report) |
Full-length transcript analyses (isoforms, alternative splicing studies, lncRNA, Phusion transcript, variant analyses, and many more) |
$171 |
$187 |
$240 |
Customers are required to submit only high-quality, DNA-free RNA extracts and provide sample quantification & QC data. Free access to web-based interactive RNA-Seq analysis platform for complimentary data analysis included. |
|
Other RNA-Seq services (SLAM-Seq, sRNA-Seq, …) |
Small RNA analysis, RNA kinetics, Poly(A) site determination |
Project-specific pricing |
Project-specific pricing |
Project-specific pricing |
Please contact ORC for more details or book a consultation. |
|
Bundle: genomic DNA extraction + Whole Genome Sequencing (end-to-end workflow from crude sample to FASTQ data) |
de novo genome assembly, GWAS, CNV studies, and many more |
$136 |
$148 |
$188 |
Low pass (lp) or standard WGS |
|
Whole Genome Sequencing (sample QC, library prep & QC, AVITI sequencing, demultiplexed FASTQ data & QC report) |
de novo genome assembly, GWAS, CNV studies, and many more |
$119 |
$130 |
$164 |
Low pass (lp) or standard WGS. Customers are required to submit high quality RNA-free DNA extracts. |
|
Bundle: Microbial DNA extraction + Metagenome-Seq (end-to-end workflow from crude sample to FASTQ data) |
De novo genome assembly, taxonomic classification, functional annotation and many more |
$154 |
$168 |
$215 |
Low pass (lp) or standard WGS. Optional host DNA removal procedure is available on demand. Please contact ORC for more details. |
|
Metagenome-Seq (sample QC, library prep & QC, AVITI sequencing, demultiplexed FASTQ data & QC report) |
De novo genome assembly, taxonomic classification, functional annotation and many more |
$119 |
$130 |
$164 |
Customers are required to submit high quality RNA-free DNA extracts. |
|
Bundle: DNA extraction + Amplicon-Seq | 16S (end-to-end workflow from crude sample to FASTQ data) |
Phylogenetic classification, environmental monitoring, diagnostics |
$134 |
$146 |
$184 |
Please contact ORC for more details. |
|
Amplicon-Seq | 16S (sample QC, library prep & QC, AVITI sequencing, demultiplexed FASTQ data & QC report) |
Phylogenetic classification, environmental monitoring, diagnostics |
$99 |
$107 |
$133 |
Customers are required to submit high quality RNA-free DNA extracts. |
|
Bundle: DNA extraction + Amplicon-Seq | Custom (end-to-end workflow from crude sample to FASTQ data) |
- |
- |
- |
- |
Will be available soon. |
|
Single cell (sc)RNA-Seq (end-to-end workflow from single cell isolation to FASTQ data) |
- |
- |
- |
- |
Will be available soon. |
|
Whole-Exome Sequencing (end-to-end workflow from crude sample to FASTQ data) |
- |
- |
- |
- |
Will be available soon. |
|
Sample plate normalization (robotic) |
Per 96-well plate |
$162 |
$167 |
$184 |
96 well plate-wise processing independent of the submitted sample number |
|
Custom SOP development |
|
Project-specific pricing |
Project-specific pricing |
Project-specific pricing |
Please contact ORC for more details or book a consultation. |
|
Instrument booking |
|
$25/booking |
$25/booking |
n/a |
First-time users are required to book a one-hour hands-on training session (see "Customer training"). Booking time cannot exceed the time to run the individual sample set. Customers are required to supply their own consumables (i.e. all reagents and labware). |
|
Customer training |
|
$80/hour |
$80/hour |
n/a |
First-time users of a bookable instrument are required to purchase this one-hour hands-on training session in addition to the instrument booking. |
Consultations
ORC offers free consultations for genomics and transcriptomics research projects. Please pick a time and book a consultation with Dr. Martin Mau (PhD), ORC Lab Manager. Click here to book a consultation time.
Booking time slots are available each Monday (1-4 p.m.) and each Friday (9 a.m.-12 p.m.). Before booking your appointment, you are required to provide:
- contact information (name and email address)
- short description of your project that you are planning to pursue in the 'Add any special requests' field
- preference for meeting (in person or through MS Teams)
Resources
For more information on sample handling and the sample submission procedure to the Omics Resource Centre review the ORC Sample Submission Guide by following the link here.
Follow this link here to a comprehensive educational beginner’s guide on how to prepare and handle samples for Next Generation Sequencing projects.
Learn more about the Element Biosciences AVITI sequencer at ORC by following the link here.
By following the link here you learn more about Next Generation Sequencing (NGS) technology and applications in the past, present and future.
More information on the principles of sequencing-by-synthesis (SBS) can be found in a Review here and in the original publications by Nyrén et al and Margulies et al.
Follow the link here to learn more about synthetic long-read (SLR) sequencing on the short-read AVITI sequencer and its advantages or disadvantages compared with other long-read sequencing technologies (e.g. Nanopore sequencing, Single-Molecule Real-Time (SMRT) sequencing).
Single-cell RNA sequencing (scRNA-seq) is a powerful genomic technique that measures gene expression in individual cells to understand cellular diversity and heterogeneity within a population. Follow the link here for a practical guide to scRNA-Seq and here for an evaluation of current (2025) commercial single-cell RNA sequencing technologies.
Learn more about GATK here – a tool designed to process exomes and whole genomes generated with NGS technology to identifying SNPs, indels, somatic short variant calling, copy number (CNV) and structural variation (SV). The Genome Analysis Toolkit (GATK) be rapidly deployed through Docker on Linux and MacOSX computing infrastructures with minimal dependencies.
Click here to enter Kbase - a community-driven research platform for systems biology. The Department of Energy Systems Biology Knowledgebase (KBase) host dozens of programs for comparative genomics, differential gene expression analysis, genome annotation, phylogenetic analysis, microbial community analysis with an easy-to-access GUI and Drag and Drop feature as well as Globus connectivity for bulk importing raw and processed sequencing data.
Learn more about Galaxy here - The open source, web-based platform is ideal for analyzing a wide array of data, authoring workflows, training and education, and publishing tools. Researchers from a Canadian institution have now access to UseGalaxy.ca, a new public Galaxy infrastructure designed to serve the Canadian research and educational community. As a Canadian researcher, you automatically get a storage quota of 500 GB and access to the support staff of the Digital Research Alliance of Canada.
Follow the link here for ExpressAnalyst - a comprehensive web-based platform for gene expression and meta-analysis data. The platform is designed for bench researchers rather than specialized bioinformaticians and integrates data processing, statistical analysis and data visualization to support 1) Data comparisons 2) Biological interpretation, and 3) Hypothesis generation. It uses either a gene list as input for knowledge-driven network analysis using or raw FASTQ files for gene expression profiling and meta-analysis.
Click here for more information on iDEP - the integrated Differential Expression & Pathway (iDEP) analysis web-based tool supports exploratory analysis of RNA-Seq data and is tailored to the needs of lab bench scientists without in-depth programming knowledge. The iDEP tool utilizes gene annotation models of over 5500 species from Ensembl and STRING-db and offers extensive tutorials and demos.
Click here to enter WebGestalt - a suite of tools for functional enrichment analysis in various biological contexts. The WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) helps researchers to explore large sets of genes and is composed of four modules: gene set management, information retrieval, organization/visualization, and statistics. Gene sets can be organized and visualized in various biological contexts, including Gene Ontology, tissue expression pattern, chromosome distribution, metabolic and signaling pathways, protein domain information and publications. Omics data can be interpreted through over-representation analysis (ORA), gene set enrichment analysis (GSEA), network topology-based analysis (NTA), metabolomics and multi-list analysis functionality.
Follow the link here to access Kangooroo – a web-based interactive platform for RNA-Seq data analysis with an intuitive GUI which allows researchers to analyze their RNA-Seq samples even without bioinformatics experience. Genome references from over 40 species are available for data analysis and additional reference species can be uploaded.
ORC customers are getting free access to Kangooroo upon purchase of an applicable RNA sequencing service. Click here to book a consultation time.
Click the link here for Cytoscape - an open-source software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression profiles and other state data. Most standard network and annotation file formats are supported by Cytoscape such as SIF (Simple Interaction Format), GML, XGMML, BioPAX, PSI-MI, GraphML, KGML (KEGG XML), SBML, OBO, and Gene Association, delimited text files and MS Excel™ Workbook. In addition, you can import data files, such as expression profiles or Gene Ontology (GO) annotations.
Click here to enter QIIME – an open-source bioinformatics software package for analyzing microbiome data, such as 16S rRNA gene sequencing and other amplicon data. The Quantitative Insights Into Microbial Ecology (QIIME) platform offers a toolkit to process, analyze, and visualize diverse microbiome data, from raw DNA sequencing to shotgun metagenomics. Its latest iteration, QIIME 2, can be utilized through graphical interfaces such as Galaxy or via command line, enabling straight-forward access on high-performance compute clusters and cloud resources.
Follow the link here to access the MIQ Score System - a fully automated microbiomics pipeline to help identify potential biases in microbiome preparation by comparing observed read abundances to expected values from a standardized microbial community with known relative abundances and manufacturing tolerances. The Measurement Integrity Quotient (MIQ) Score System is available as web-based platform through an online portal or designed to be rapidly deployed through Docker across different computing infrastructures to analyze either 16S Sequencing or Shotgun Metagenomic Sequencing.
Click the link here to access MultiQC - a reporting tool that parses results and statistics from bioinformatics tool outputs, such as log files and console outputs. It helps to summarize experiments containing multiple samples and multiple analysis steps, such as examining NGS sequencing irregularities across large sample sets and dataset filtering in single cell data and population studies.
Click here to access GenomeQC – an assessment tool for genome assemblies and gene structure annotations. The tool provides a complete and quick Benchmarking of Universal Single-Copy Orthologs (BUSCO) analysis as well as standard assembly metrices (L50, N50, LG50, etc.).
Follow the links to JBrowse here and IGV here – two of the most widely used genome browsers to visualize common genome analysis data types such as BAM, GFF, and BigWig, in addition to Hi-C data rendering and UCSC track hubs. Both browsers are available as desktop, web and command line application.
Click this link to access the JGI Data Portal here and the IMG here – Both the Joint Genome Institute (JGI) Data Portal and the Integrated Microbial Genomes (IMG) system serve as a community resource for analysis and annotation of plant, algal, fungal, and microbial genome and metagenome datasets in a comprehensive comparative context. You can explore the dataset manually in your browser or download the data to your server or local machine via Globus, command line, browser or API.
For more information on software tools, databases and services for bioinformatics and life sciences follow the link here to visit bio.tools - a community-driven registry of bioinformatics software and data resources.
Follow the link her to a enter iBiology:Techniques – a collection of seminars on a variety of techniques from the principles of next generation sequencing to bio imaging.
Click her to access JoVE - a peer-reviewed scientific journal that publishes experimental methods in video format in aeras such as genetics, biology, developmental biology and others.
FAQs
Have a question? Check out our FAQ section to view our responses to frequently asked questions.
Sample submissions
New customers must initially contact orc@usask.ca to request a quote. Ideally, this request happens after the customer had booked a free-of-charge consultation with the ORC lab manager in person or online via Teams. Click here to book a consultation.
After acceptance of the quote in writing, the customer will return the completed ORC Sample Submission Template providing their contact and financial information and any required information related to their project and samples. For more information on the sample submission procedure follow the ORC Sample Submission Guide.
The ORC Lab Manager will review the service request and permit the sample submission based on the completeness of the information given.
We are currently implementing a Customer Portal which which will substitute the current ORC Sample Submission Template for sample submissions in the future.
You can submit any solid or liquid crude sample and FFPE sample for nucleic acid (NA) extraction and downstream processing (e.g. sample QC, NGS library preparation, sequencing).
Alternatively, you can also submit any nucleic acid samples (cDNA, RNA, DNA) and library pools for processing and/or direct sequencing.
Click here to view ORC’s Sample Submission Guideline and follow the guide's detailed instructions.
For samples that are new to our workflows, we may ask you to submit a few test samples (if available) in advance of your project sample submission.
We strongly recommend submitting crude samples in either 2 ml DNA microcentrifuge tubes with Safe-Lock or Snap-Lock design (e.g. Eppendorf or Axygen) or in 96-deep-well plates (e.g. Greiner Bio-One Masterblock® 96-Well 1ml Deep Well).
For nucleic acid extracts or library pools, we require their submission in nuclease-free sterile 1.5 ml low-binding DNA microcentrifuge tubes with Safe-Lock or Snap-Lock design (e.g. Eppendorf or Axygen) or in standard skirted or non-skirted 96-well PCR plates.
When submitting 96-well plates, they need to be covered by an adhesive seal when stored in standard -20 C freezers and fridges (e.g. Thermo Scientific™ Adhesive PCR Plate Seal, cat. no. AB-0558), or they need to be thermo-sealed when stored at -80 C or in liquid nitrogen (e.g. LGC Easy-peel well plate seal).
We require printed labels or legible handwriting that includes a running integer per sample (e.g. 1, 2, 3…n) on the top and on the side of a tube in addition to the ORC Estimate ID (e.g. JOHN_DOE_WCVM-ORC-25-0521-3) on the tube box as per sample submission documentation. Alternatively, you can submit samples in 96-well plates which are required to be labelled with the ORC Estimate ID on the top and side.
Detailed information can be found in the ORC Customer Portal, the ORC Sample Submission Template, and the ORC Sample Submission Guideline.
If you cannot transfer your samples into the requested sample submission formats (i.e. larger volumes or amounts and/or other sample storage container formats) ORC will add project-specific surcharges for extended sample handling to your estimate. Please click here to view ORC’s Sample Submission Guideline.
Yes, ideally you accompany your sample submission with a gel image or Bioanalyzer/Tapestation traces and information. Please provide this information through the ORC Customer Portal or by email to orc@usask.ca accompanied with the completed ORC Sample Submission Template.
If you do not accompany QC data with your processed samples (i.e. NA extract or NGS libraries), ORC will provide the QC service for you and will add these costs to your estimate.
Yes, ORC offers a direct sequencing service that includes the QC and quantification of your library pool and the sequencing on our AVITI sequencer. We do prefer the submission of library pools over submission of individual libraries. Library pools must be submitted in nuclease-free sterile 1.5 ml low-binding DNA microcentrifuge tubes with Safe-Lock or Snap-Lock design (e.g. Eppendorf or Axygen).
You are required to accompany your samples with an Excel file containing the index and adapter information through the ORC Customer Portal or together with the ORC Sample Submission Template to orc@usask.ca.
Data handling
The authorized email for data access is the email address used to place an order through the ORC Customer Portal or the ORC Sample Submission Template. Upon order completion, you will receive a data release email with download instructions. ORC’s standard process for all on-campus USask customers and most external academic/non-profit institutional facilities is data delivery through the Globus platform using your institutional account and sign-on credentials.
For non-institutional external customers, ORC shares the data as a Globus Guest Collection. ORC creates a special folder on its Globus endpoint, grants the customer specific read permissions, and sends the customer a link to access that folder directly through the Globus web app. The customer will then be prompted to authenticate using a temporary, one-time identity to download the files (follow the link here for Detailed Instructions).
In case the standard process for data delivery does not work for you, ORC may be able to deliver the data directly to your AWS S3 bucket or any other endpoint on your system. Or, as a last resort, you can choose to have data shipped on a hard drive purchased by ORC and invoiced to the customer at a cost of C$300 per hard drive.
If you encounter any problems downloading your data, please report the issue in a reply to the data release email.
If you get the "Permission denied" error, it means that your current account does not have access to this folder. Please go to "Settings" (tab on the left side) and see what is reported as the "Identity." This error indicates a mismatch between the login email/account and shared email/account. To remedy the issue, follow these steps:
- From the ‘Settings’ page, select "Link another identity" found on the top right of the screen
- Follow the prompts and select your institution from the list. Or, if you cannot find it, select the “You can also link an email address to your Globus account” link.
- Link your current account to the email address you would like to use and follow the confirmation steps.
- Retry accessing the shared collection.
If you are still having trouble, please email orc@usask.ca and request that the reported email/account be added to your folder for your project (please list the ORC Estimate ID/quote number).
If you can access the collection but the page will not load and instead reports “Retrieving directory contents” for an unusual amount of time, it is possible that the content is being blocked by your institutions firewall. This appears to be a common issue for some institutional partners.
Please contact your IT team to allow Globus traffic on your network. As well, please contact orc@usask.ca to report this issue and get a fast resolution.
ORC will deliver the order results in FASTQ format (raw reads and quality scores), unless other formats are requested on the quote and the customer agrees to pay additional charges, if any.
In addition, the sequencing orders will include a methods document and a summary of some overall sequencing statistics for each sample. Optionally, the customer can request the adapter sequences to be trimmed free of charge.
We do not archive sequencing data. ORC holds the data and samples for a limited timeframe only (maximum of two months from the release email) and recommends downloading or transferring the data at your earliest convenience after receiving the email notification.
If you wish to claim your libraries or leftover crude samples and extracts (if available), please state this in a reply to the data release email. After the timeframe noted above, the customer’s data will be deleted without further notice to the customer.
While ORC does not currently offer data analysis services, we do plan to offer this service in the future. Nonetheless, we are happy to help get you started with your analysis through guidance in analysis software selection.
For example, if you are looking for a bioinformatic program with a graphical interface, Galaxy and Geneious are two popular and powerful tools with plenty of documentation. KBase is also a great cloud-based platform that hosts many common command-line packages in point-and-click format along with a plethora of premade pipelines and tutorials.
For more information, visit the ORC resources section or visit the ORC consultation section to book a session.
General topics
Currently, ORC does not offer long-read sequencing services. For more information on our current services, view ORC's Services and Pricing sections or click here to book a consultation.
No, the booking equipment is limited to the following instruments:
- Agilent Tapestation 4200
- Agilent Synergy LX Microplate reader
- KingFisher Apex Nucleic Acid Purification System
- Invitrogen™ Countess™ 3 FL Automated Cell Counter
- Miltenyi Biotec gentleMACS™ Dissociator
ORC services are co-ordinated based on project size, urgency and staff availability. Standard turnaround times vary, but here are the typical tiime ranges:
- two to three weeks for end-to-end services (i.e. crude sample prep + library prep + sequencing)
- one to two weeks for sequencing only (libraries already prepared)
- one to two days for instrument booking or short consultations
ORC will be closed for specific holidays, and we will be unable to accept packages during those days. Additionally, the closures will offset the typical turnaround times for orders. Please plan ahead when shipping your samples.
ORC closures:
- National Day for Truth and Reconciliation (Sept. 30)
- Saskatchewan public holidays
- USask winter break: December 23, 2025, to January 4, 2026
ORC provides access to complex genomic technologies and offers guidance in experimental design. One measure of our impact on the USask research community is the number of publications that ORC supports. ORC requests that any data generated using ORC resources (presented in a publication, grant proposal, poster or presentation) be formally acknowledged.
For more details on the best format for acknowledgment, review ORC's Terms and Conditions.
Contact Us

ORC hours of operation: Monday to Friday, 9 a.m. to 4:30 p.m.
Click here to access ORC's terms and conditions.
Questions or suggestions? Contact:
Dr. Martin Mau
Genomics Laboratory Manager
Western College of Veterinary Medicine
University of Saskatchewan
52 Campus Dr, Saskatoon, SK S7N 5B4
orc@usask.ca
Lab: WCVM 2207 | Office: WCVM 2209
Thank you to our funders


